UK lighthouse laboratories testing for SARS-COV-2 may have breached WHO Emergency Use Assessment and potentially violated Manufacturer Instructions for Use
Fri 10:25 am +01:00, 26 Feb 2021 
If you have been following the coverage on this blog about Covid-19 false positives you will be interested to read that we have recently discovered that UK laboratories have been routinely recording a large proportion of Covid-19 test results as positive based on the presence of one target gene alone, when there should have been two or more, as required to comply with WHO rules and manufacturer instructions. For example, an average of 35% across the whole of the UK during week of 25 Jan 2021 were based on one gene only. Obviously, there is higher risk of encountering false positives when testing for single genes alone, because of the possibility of cross-reactivity with other HCOVs and prevalent nasopharyngeal bacteria or reagent contamination. Unless UK laboratories have performed diagnostic validation of their single gene call, for both the original and the B1.1.7 variant, and there is no evidence of this in the public domain, it can only be assumed that, in the absence of confirmatory testing, many of the reported positive results may in fact be inconclusive, negative or from people who suffered past infection for SARS-COV-2.
An academic pre-print of article available here
Positive results from UK single gene testing for SARS-COV-2 may be inconclusive, negative, or detecting past infections
Prof. Martin Neil, School of Electronic Engineering and Computer Science, Queen Mary, University of London
25 February 2021 (version 2)
Abstract
The UK Office for National Statistics (ONS) publish a regular infection survey that reports data on positive RT-PCR test results for SARS-COV-2 virus. This survey reports that a large proportion of positive test results are based on the detection of a single gene rather than on two or more genes as required in the manufacturer instructions for use, and by the WHO in their emergency use assessment. The proportion of positives called on single genes increased from mid-November to mid-December 2020, suggesting a shift in testing policy coincident with the reported significant increase in transmission of the new variant B1.1.7, and again starting January 2021. Without diagnostic validation of the single gene call, for both the original and the B1.1.7 variant it can only be assumed that, in the absence of confirmatory testing, many of the reported positive results may in fact be inconclusive, negative or from people who suffered past infection for SARS-COV-2.
Background
The ONS publish a regular infection survey [1] that includes data from two UK lighthouse laboratories, based in Glasgow and Milton Keynes, where both use the same RT-PCR test kit, to detect the SARS-COV-2 virus. This survey includes data on the cycle threshold (Ct) used to detect positive samples, the percentage of positive test results arising from using RT-PCR, and the combinations of the SARS-COV-2 virus genes tested that gave rise to positives between 21 September 2020 and 30 January 2021 across the whole of the UK.
ThermoFisher TaqPath kit[1] is used by the Glasgow and Milton Keynes lighthouse laboratories to test for the presence of three genes from SARS-COV-2[2]. Despite Corman et al [2] originating the use of PCR testing for SARS-COV-2 genes[3] there is no agreed international standard for SARS-COV-2 testing. Instead, the World Health Organisation (WHO) leaves it up to the manufacturer to determine what genes to use and merely requires end users to adhere to the manufacturer instructions for use (IFU). As a result of this we now have an opaque plethora of commercially available testing kits, that can be applied using a variety of test criteria. Other UK laboratories use different testing kit, and test for different genes.
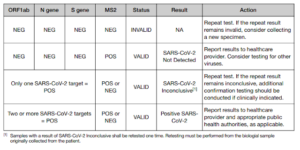
The WHO’s emergency use assessment (EUA) for the ThermoFisher TaqPath kit [3], used by the Glasgow and Milton Keynes lighthouse laboratories, includes the instruction manual and contained therein is an interpretation algorithm describing an unequivocal requirement that two-or-more genes be detected before a positive result can be declared. The WHO have been so concerned about correct use of RT-PCR kit that on 20 January 2021 they issued a notice for PCR users imploring them to review manufacturer IFUs carefully and adhere to them fully [4].
Increasing proportion of single gene “calls”
The ONS’s report [1] lists SARS-COV-2 positive results for valid two and three gene combinations[4] from the Glasgow and Milton Keynes lighthouse laboratories. However, it also lists inconclusive single gene detections as positive results[5]. This use of single gene “calls” therefore suggests that Glasgow and Milton Keynes lighthouse laboratories may have breached WHO emergency use assessment (EUA) and may have violated the manufacturer instructions for use (IFU). Indeed, Section 10 of this ONS Covid-19 Infection survey on the 8 January 2020 [16] stated that (emphasis mine):
“Swabs are tested for three genes present in the coronavirus: N protein, S protein and ORF1ab. Each swab can have any one, any two or all three genes detected. Positives are those where one or more of these genes is detected in the swab …..”
Over the period reported the maximum percentage of positives on a single gene is 35% for the whole of the UK for the week of 25 January. The overall UK average is 21%. The maximum percentage reported is 65%, in East England in the week beginning 5 October. In Wales it is 48%, in Northern Ireland it is 47% and in Scotland it is 49%. The full data including averages and maxima/minima are given in Table 1.
Figures 1 and 2 show the percentage of weekly single gene positives across the UK nations and English regions. There was a significant peak across the whole of the UK (except NI) from mid-November to early December, coincident with the reported significant increase in transmission of the new variant B1.1.7 [5]. However, given this new variant was concentrated in Kent and NE London, with limited spread into the rest of London, Anglia and Essex its presence cannot explain why single gene calls increased in other regions such as the North East and Yorks. & Humber. There has been another significant increase in the percentage of single gene positives since the end of 2020, rising throughout January, and here the rise is steady across all English regions and UK nations.
In startling contradiction to the ONS reports Professor Alan McNally, Director of the University of Birmingham Turnkey laboratory, who helped set up the Milton Keynes lighthouse laboratory, reported in the Guardian newspaper, in an article about the new variant, that all lighthouse laboratories operated a policy that adhered to the manufacturer instructions for use: requiring two-or-more genes for positive detection [6] (this policy is also documented in the supplementary material provided in [7]).
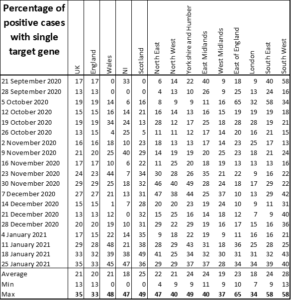
Table 1: Percentage of weekly single gene positives from 21 September 2020 to 25 January 2021, including averages and maxima/minima
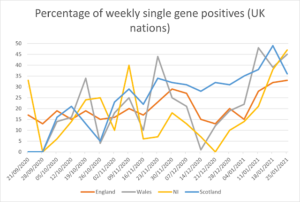
Figure 1: Percentage of weekly single gene positives from 21 September 2020 to 25 January 2021 (UK nations)
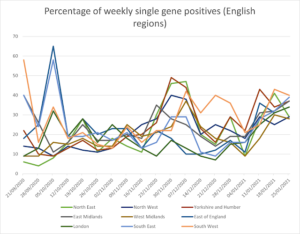
Figure 2: Percentage of weekly single gene positives from 21 September 2020 to 25 January 2021 (English regions)
In April 2020, the UK lighthouse laboratories were testing for single genes and discounted the S gene as early as mid-May, months before the discovery of the new variant B1.1.7 (emphasis mine):
“Swabs were analysed at the UK’s national Lighthouse Laboratories at Milton Keynes (National Biocentre) (from 26 April) and Glasgow (from 16 August) …., with swabs from specific regions sent consistently to one laboratory. RT-PCR for three SARS-CoV-2 genes (N protein, S protein and ORF1ab) ….. Samples are called positive in the presence of at least single N gene and/or ORF1ab but may be accompanied with S gene (1, 2 or 3 gene positives). S gene is not considered a reliable single gene positive (as of mid-May 2020).”
Indeed, in Table 1 of [17] 18% of tests were positive on one gene only and it was concluded, in Table 2 of [17], that for people with single gene positives, when Ct > 34, none had symptoms and for people with Ct < 34 only 33% had symptoms.
Furthermore in [16], published January 8th 2021, it states the goal of using one gene was explicitly to approximate the growth of the new variant (emphasis mine):
“There has recently been an increase in the percentage of positive cases where only the ORF1ab- and N-genes were found and a decrease in the percentage of cases with all three genes. We can use this information to approximate the growth of the new variant.”
Quality control and cross reactivity
Quality control problems have already been reported in UK laboratories [8, 9, 10] and there have been concerns expressed about the potential for false positives arising consequently. Recent suspicion focused on problems potentially caused by breaches in acceptable Ct thresholds (Ct > 37), suggesting no, or past, infection. However, this new ONS data shows there may be an additional potentially dominant source of false positives, at least within the period covered by the ONS report, if not from April 2020; specifically, positives caused by potential breach of WHO end user assessment and manufacturer instructions for use. These sources of false positives appear not to be caused by statistical error but might instead be categorised as systematic non-compliance.
Concerns about testing in commercial laboratories were documented by the ONS as early as May 2020 [11], when the REACT study discovered that circa 40% of positive tests from commercial laboratories were in fact false positives. A similar false positive rate (44%) was reported in Australia [12] in April 2020. More recently Nicholas Lewis claims that, despite very low false positive rates (0.033%) from testing done by non-commercial and academic laboratories, there may be good reason to suspect the operational false positive rates from lighthouse laboratories are worse than these by some orders of magnitude [13].
Obviously, there is higher risk of encountering false positives when testing for single genes alone, because of the possibility of cross-reactivity with other HCOVs and prevalent nasopharyngeal bacteria or reagent contamination. The potential for cross reactivity when testing for SARS-COV-2 has already been confirmed by the German Instand laboratory report from April 2020 [14]. This report describes the systematic blind testing of positive and negative samples anonymously sent to many laboratories throughout Germany and evaluated for the presence of a variety of genes associated with SARS-COV-2[6]. They reported significant cross reactivity and resultant false positives for OC43, and HCoV 229E (a common cold virus) as well as for SARS-COV-2 negative samples, not containing any competing pathogen.
It should be noted that these issues are not unique to the UK. Likewise, 70 Dutch laboratories were surveyed in November 2020 [15], by the National Institute for Public Health and the Environment, with 76 diagnostic workflows reported as using only one target gene to diagnose the presence of SARS-COV-2 (46% of all workflows).
Conclusions
Unless the UK lighthouse laboratories have performed diagnostic validation of their single gene call, for both the original and the B1.1.7 variant, and there is no evidence of this in the public domain, it can only be assumed that, in the absence of confirmatory testing, many of the reported positive results may be inconclusive, negative or from people who suffered past infection for SARS-COV-2. Even with diagnostic validation of the single gene call, the UK lighthouse laboratories appear to be in breach of both the WHO emergency use assessment and, also, to have potentially violated the ThermoFisher TaqPath kit instructions for use.
http://probabilityandlaw.blogspot.com/2021/02/uk-lighthouse-laboratories-testing-for.html


